
Bicubic spline is a fast and precise two-dimensional interpolation and fitting method. ALGLIB package contains an implementation of 2D splines available in several programming languages:
Our implementation of 2D splines:
1 Getting started and examples
Getting started
Fitting splines to scattered data
2 Benchmarks
Medium-scale problems: bicubic spline fitting vs RBFs
Large-scale problems: BlockLLS vs FastDDM
3 Downloads section
Bicubic spline interpolation functionality is provided by the spline2d subpackage of ALGLIB package. A bicubic spline can be created from the data sampled at the regular grid (to be exact, more general rectilinear one) with spline2dbuildbicubicv function. This function supports both scalar and vector-valued splines.
After an instance of spline object is built, you can perform following operations:
ALGLIB Reference Manual includes following examples which show how to work with bicubic splines:
In the previous section we mentioned that bicubic spline needs data to be sampled at the regular grid. The reason is that internally bicubic spline is composed of many rectangular cubic patches, so there must be a one-to-one correspondence between spline nodes and dataset points. Such one-to-one correspondence allows fast and memory-efficient construction of the bicubic spline from the array of spline values.
However, it is also possible to fit bicubic spline in the least squares sense to the irregular (scattered) data. Efficient utilization of the problem sparsity allows us to solve really large tasks - to utilize grids from 100x100 to 500x500 nodes or even (with FastDDM algorithm) with 10.000x10.000 nodes.
ALGLIB has two least squares solvers for bicubic splines:
An example of bicubic spline fitting - spline2d_fit_blocklls - can be found in ALGLIB Reference Manual.
Radial basis function interpolation is a well-known method of handling scattered multidimensional data. ALGLIB includes sophisticated implementation of RBFs called HRBF (Hierarchical RBF). Our implementation of RBFs uses many algorithmic improvements and is much faster than straightforward attempt to fit RBFs by solving huge dense linear systems. However, in the 2D case bicubic splines become a viable alternative with an order of magnitude improvement in both memory consumption and running time.
Our first test compares ALGLIB multilayered RBFs vs. BlockLLS and FastDDM bicubic solvers. We tested running times of these methods on a 96x96 grid being fitted to 20K randomly generated points. A hierarchical RBF model was configured to fit three successive layers with initial radius set to 1.0. Such problem can be classified as medium scale one (roughly 10.000 parameters to fit). A 4-core 3.5 GHz x64 CPU running C++ version of the ALGLIB (Intel MKL included) was used.
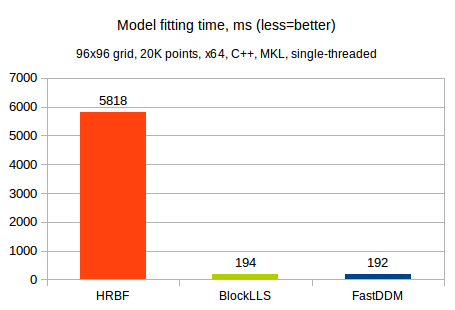
You can see that both bicubic fitting algorithms (BlockLLS and FastDDM) were much faster than HRBF.
The main difference between BlockLLS and FastDDM solvers is that former solves entire problem in one step and latter uses recursive divide-and-conquer approach.
The BlockLLS method uses sparse LSQR linear solver with block septa-diagonal preconditioner matrix (hence the method's name). The linear systems being solved are extremely sparse but nevertheless, for grids larger than 512x512 even highly optimized BlockLLS becomes too memory-hungry. The FastDDM algorithm solves this problem by splitting large tasks into smaller chunks and handling them with BlockLLS-like "basecase" algorithm.
We tested running times and memory consumption of both methods on a 512x512 grid being fitted to 525K randomly generated points. The same CPU as in the previous test was used. Below we compare running times of single threaded BlockLLS and FastDDM solvers:
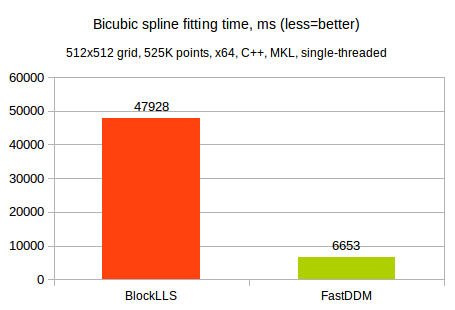
Well, we have to say that both solvers show impressive results. A 512x512 grid means that we have 262.000 variables to fit. Being able to solve such huge least squares problem in just 48 seconds (BlockLLS time) is impressive anyway. And 7 seconds spent by FastDDM are even more impressive.
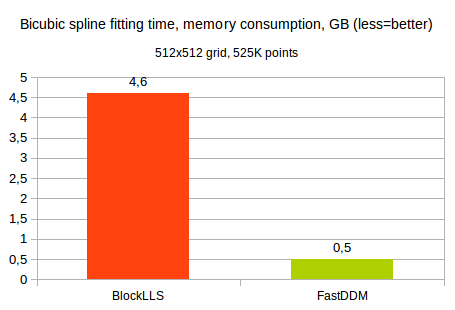
Just for comparison, a brute-force attempt to use dense Cholesky solver would need about 150.000-200.000 seconds (roughly 2 days!) and about 500 GB of memory. Contrary to that, BlockLLS needs roughly 4.6 GB of memory (actual memory consumption according to OS stats), and FastDDM needs just 0.5 GB:
This article is licensed for personal use only.
ALGLIB Project offers you two editions of ALGLIB:
ALGLIB Free Edition:
+delivered for free
+offers full set of numerical functionality
+extensive algorithmic optimizations
-no multithreading
-non-commercial license
ALGLIB Commercial Edition:
+flexible pricing
+offers full set of numerical functionality
+extensive algorithmic optimizations
+high performance (SMP, SIMD)
+commercial license with support plan
Links to download sections for Free and Commercial editions can be found below:




